Work Packages
We have designed 5 work packages in which psychiatrists and pathologists (WP1), cell biologists with extensive experience in omics-technologies (WP2), and computational scientists (WP3) will closely collaborate with chemists (WP4) and neurobiologists (WP5).
Donor recruitment and phenotyping
Leaders: Jörg Hamann en Karel Scheepstra
Work Package 1 has the aim to sustain and complement the long-term psychiatric brain donor program of the Netherlands Brain Bank (NBB) to collect brain tissue from well-characterized donors to facilitate fundamental and translational psychiatric research. The collection of high-quality, deeply phenotyped brain material from patients with neuropsychiatric disorders and control subjects is important for the international research community and Dutch researchers, including the iCNS consortium.
WP1 builds on the successful NBB-Psy donor program, focused on the collection of brain samples and data from donors diagnosed with major depression (MDD), schizophrenia (SCZ), bipolar disorder (BD), obsessive-compulsive disorder (OCD), post-traumatic stress disorder (PTSD), autistic spectrum disorder (ASD), attention-deficit hyperactivity disorder (ADHD), and controls. We will strengthen and expand this existing, national infrastructure with a biobank collecting (cerebrospinal fluid) CSF and blood from patients with psychiatric disorders. The activities of WP1 will enable molecular psychiatric research within iCNS and future open-access dissemination to researchers globally. iCNS will set new standards in brain banking by moving beyond clinical diagnoses and incorporate clinical and pathological heterogeneity into standardized assessments.
Molecular human brain atlas
Leaders: Lot de Witte and Bart Eggen
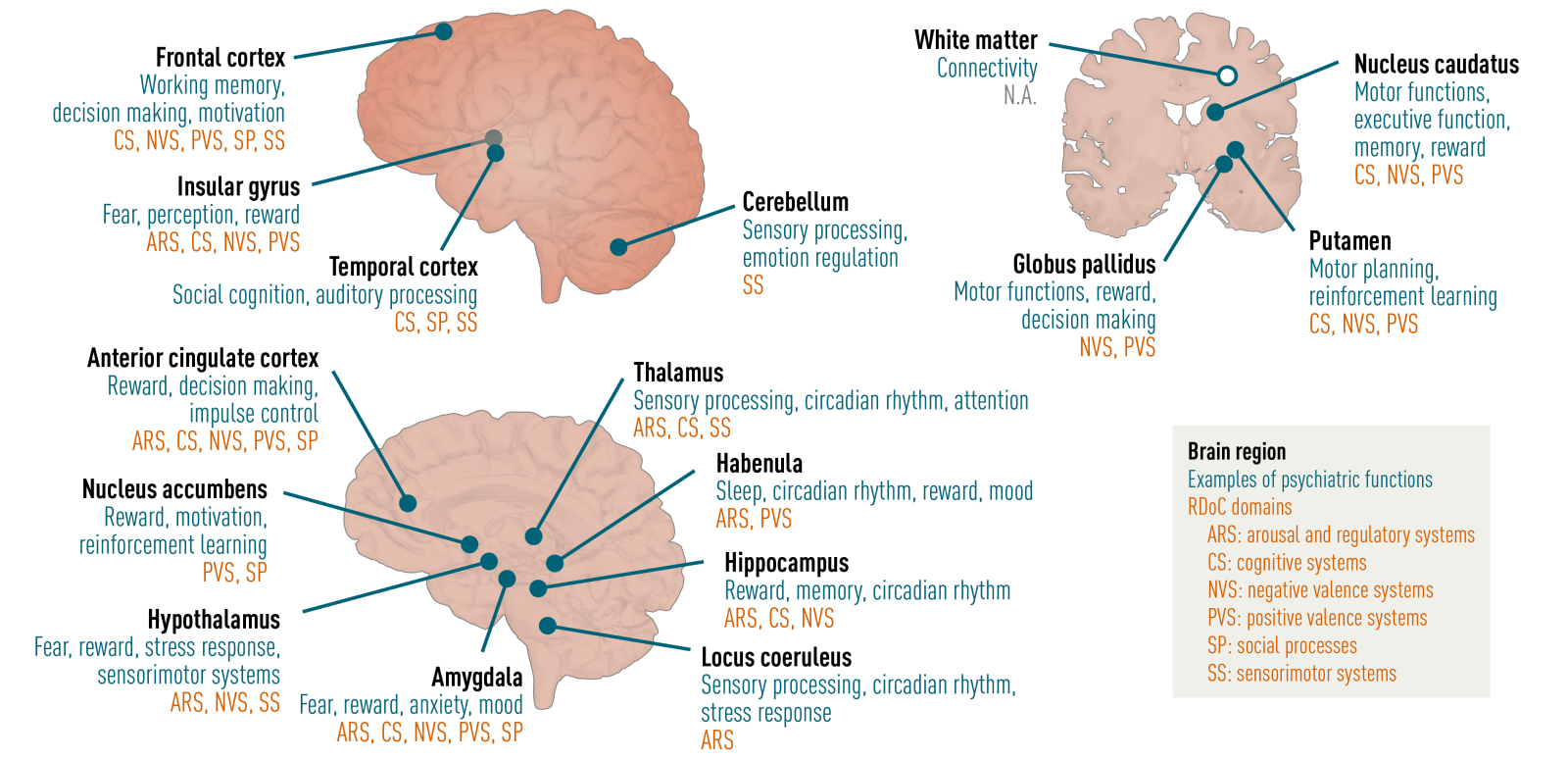
The aim of Work Package 2 is to create a molecular human brain atlas with a focus on brain regions associated with psychiatric disorders by means of a multi-omics approach. These include spatial transcriptomics, chemical proteomics and metabolomics, at single cell resolution of brain regions across multiple neuropsychiatric disorders.
The advent of several revolutionary and powerful new technologies in molecular biology and chemistry, likesingle cell transcriptomics, high resolution spatial transcriptomics, (chemical) proteomics and advanced microscopy methods, allows us to study not only potential changes in gene activity, but also the spatiotemporalfunction of proteins in postmortem human brain at the nanoscale. Using these tools, we aim to construct a molecular atlas of the human brain at the subcellular scale with the intent to delineate the molecular processes that underlie the network and cell-cell interaction changes in psychiatric disorders.
We will generate a multi-region molecular atlas of NBB donors with extensive clinical phenotypes available and the deep phenotyping of molecular and cellular features associated with selected phenotypes in specific brainregions. We will zoom in on neuron-glia interactions to delineate aberrant cell-cell interactions and communication in psychiatric disorders. The targets identified will be validated in independent datasets andcharacterized at a molecular and cellular level and across regions – patients – tissue types.
Target identification using the Netherlands Neurogenomics Database (NND)
Leaders: Inge Holtman and Ahmed Mahfouz
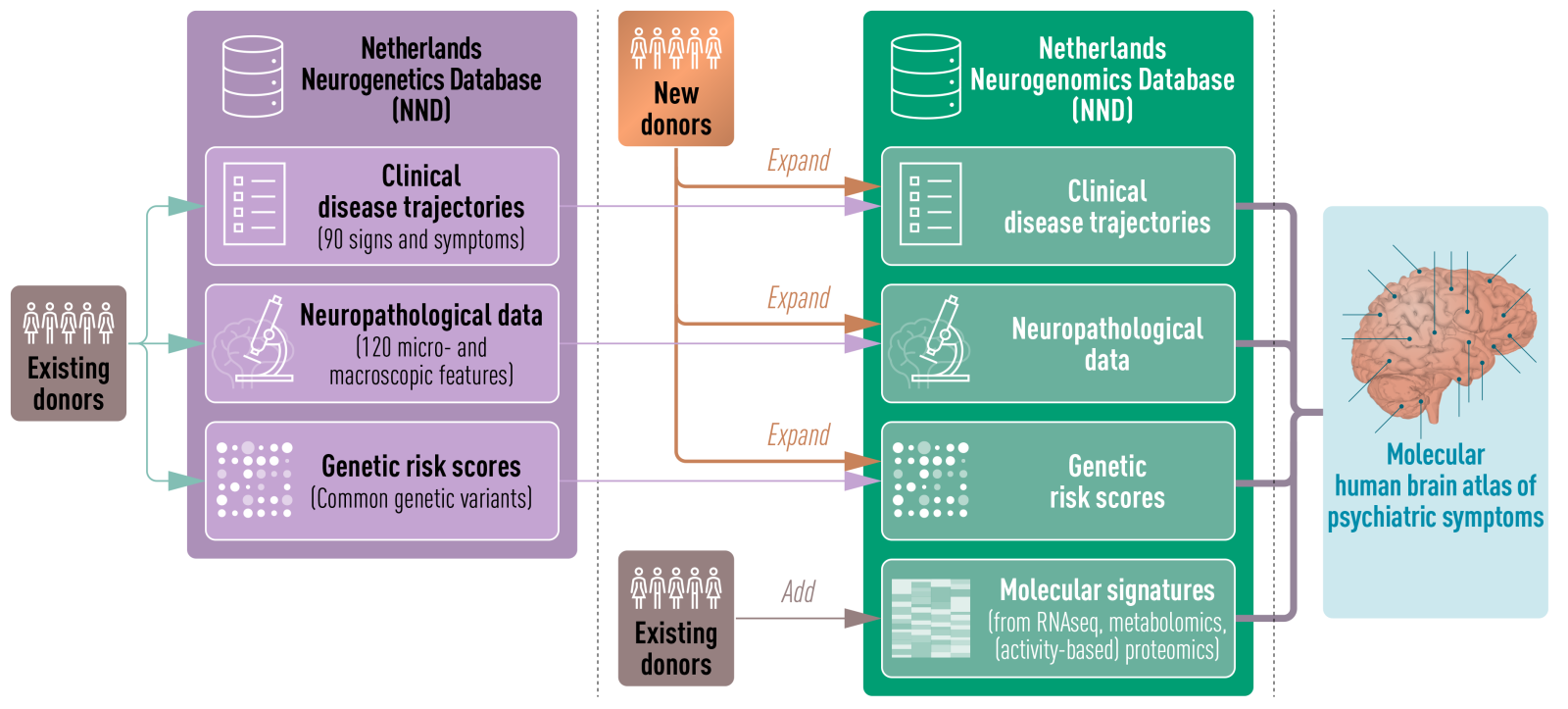
Work Package 3 aims to apply state-of-the-art statistical and machine learning approaches to integrate a wide range of multi-modal data to better understand the neurobiology of neuropsychiatric disorders, and identify novel targets. Members of the iCNS consortium will be generating large collections of multi-modal datasets (ranging from genetic risk scores, genomics profiles, chemical biology data, and clinical disease trajectories) from brain donors of the Netherlands Brain Bank. The Netherlands Neurogenomics Database (NND) will be used to track samples, and datasets to facilitate multi-modal data integration, and allow for manual and data driven curation. WP3 will integrate these datasets with multi-modal datasets from other consortia such as PsychEncode, with the ultimate aim of identifying neurobiological mechanisms of disease, and critical cell type interaction networks driving disease progression. Data driven predicted targets will be investigated in other WP of the iCNS consortium in more detail.
The NND database will also serve as a framework for the predictive modeling of neuropathologies, identifying molecular and clinical endophenotypes to enhance diagnostic accuracy. Additionally, WP3 will enable the unbiased analysis of neuron-glia interactions across multiple brain disorders using advanced computational methods. By offering open access, the NND aims to become an important resource for biomedical researchers worldwide, for innovative, integrative analysis of complex neurogenomic data across disciplines.
The NND builds on recent advances in neurogenetics and aims to create a robust and integrative resource, central to the iCNS project’s goals of elucidating molecular and cellular mechanisms underlying brain disorders.
Development of chemical probes and methods for the molecular human brain atlas and target validation
Leader: Sander van Kasteren
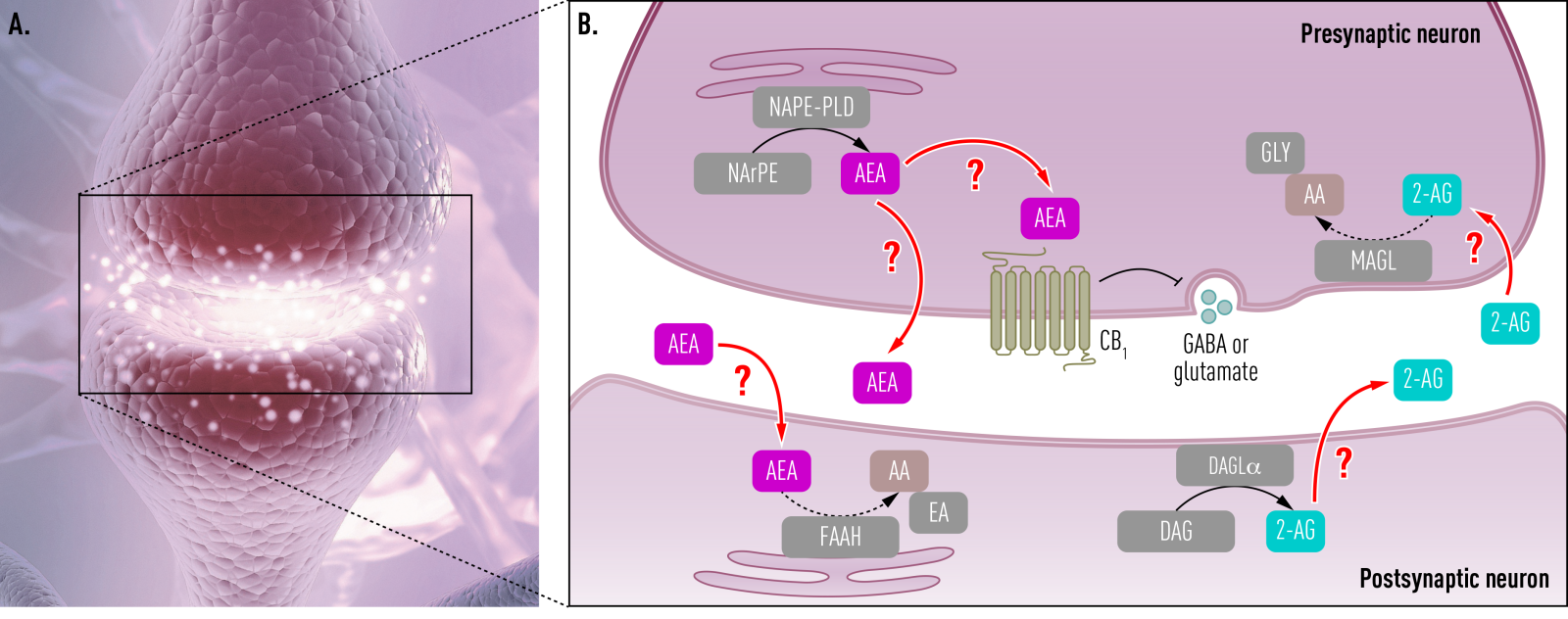
Work Package 4 aims to provide new chemical approaches that will allow the study of the brain samples beyond DNA and RNA. It will consist of work packages for the spatial analysis and visualisation of certain enzyme activities in the brain, such as those involved in the production and destruction of neuro-active ligands. The reason being that enzyme activities and mRNA levels for these proteins correlate very poorly in the brain and therefore this actual activity information is needed on how enzymes affect function.
WP4 also aims to develop tools to manipulate protein levels in the primary human brain. The challenge here is that it is primary human tissue, which means that knockout technology is not compatible with this approach. Instead, we will therefore develop new chemical mutagenesis and knockout methods. Moreover, new methods and reagents will be developed to see cells interacting in the brain. One of the key hypothesis of the proposal is that the interaction of different cell types in the brain is deregulated leading to disease. The study of this phenomenon requires new approaches to image these interactions down to the atomic level, which we will develop here. The final set of new methods developed is to image the flows of nutrients and other small molecules through the brain. This will help understand some of the other ways in which the different cells in the brain influence each other.
In order to successfully implement these new technologies in service of the questions the proposal aims to ask, the researchers in this WP will work closely with the other WPs to ensure that the reagents are all compatible with the experimental systems and models used in the wider proposal. All projects will therefore be twinned with projects from biological researchers.
Target validation in advanced model systems
Leaders: Aniko Korosi & Femke de Vrij
Work Package 5 aims to provide a mechanistic, causal and functional understanding of target molecules in relation to psychiatric symptomatology. Researchers of this WP will develop a research pipeline to study neuron-glia interactions in vitro and in vivo to provide mechanistic, causal and functional understanding of target molecules in relation to psychiatric symptomatology. WP5 will identify and test target molecules, currently known ones, or newly identified ones from WP2 and WP3, for innovative therapeutics to ultimately reverse dysfunctions in neuropsychiatric disorders.
To achieve these goals, researchers in WP5 will closely collaborate with chemists in WP4, to leverage state-of-the-art chemical tools and/or develop novel viral tools for visualization. New chemical probes may allow visualization of disease-relevant enzymes at the required high spatial resolution to build a better understanding of the nanoscale architecture of synapse dysfunction. To characterize and target neuron-glia interactions in a human context, researchers will take advantage of platforms, such as human induced pluripotent stem cell (hiPSC)-derived, neuron-glia cultures and brain organoid models.
To understand causality, cellular specificity as well as links to protein dysfunction and changes in behavior and cognition, the research in WP5 will be performed along hierarchical levels of complexity, ranging from cellular electrophysiological studies, in neural networks in vitro, to behavioral screening in in vivo animal models. The ultimate goal of this WP is to better understand causality, cellular specificity, and the connections between protein dysfunction and changes in behavior and cognition in neuropsychiatric disorders.
.jpg)
